Research Interests |
|

 https://orcid.org/0000-0002-7173-0372
https://orcid.org/0000-0002-7173-0372My research interests are in the structure and function of a wide range of important biological macromolecules, including:
I. Chromatin structure remodeling mechanism in yeast
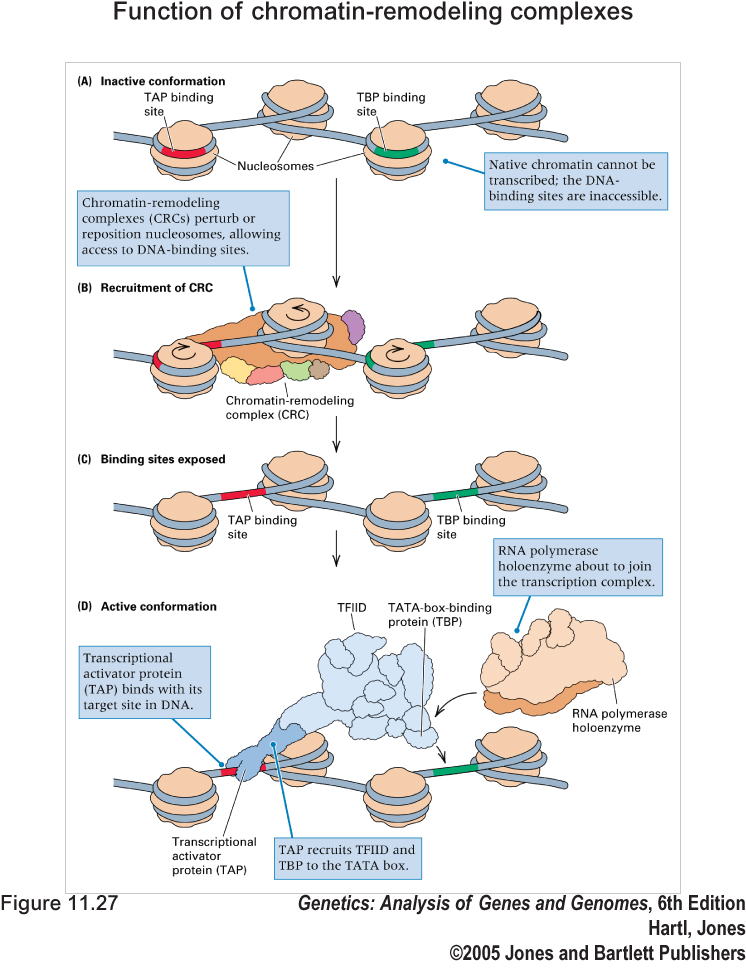
The eukaryotic genome is packaged into chromatin, which can inhibit the accessibility of certain DNA binding factors to their cognate sites in vivo. However, changes in chromatin structure play an important role in processes such as transcription, replication, DNA damage repair, and recombination. Two such ATP-dependent chromatin remodeling complexes in yeast Saccharomyces cerevisiae include the SWI/SNF complex and the RSC complex (Remodels the Structure of Chromatin). Of the two, only RSC is essential for viability, serving an important function in cell cycle progression from G2 to M. RSC is an abundant complex of at least 16 subunits, which was purified on the basis of homology of several of its subunits to those of the SWI/SNF complex.
Function of chromatin-remodeling complexes. (A) Native chromatin may conceal key DNA-binding sites. (B) A chromatin-remodeling complex either repositions the nucleosomes along the DNA or chemically modifies the histones. (C) DNA-binding sites become accessible. (D) The transcription complex is recruited to the site.
This diagram is adapted from Genetics: Analysis of Genes and Genomes. 6th Ed., Chap.11 (2005) [Publisher]
II. JAK/STAT signaling pathway in fruitfly
The signal transduction pathway conducted by JAK (Janus Activated Kinases) and STAT(Signal Transducers and Activators of Transcription) plays a critical role in hematopoiesis and immune response, and mis-regulation of the pathways is associated with a wide range of immunologic diseases and to leukemia in humans. However, the structure/function relationship of the interaction between cytokine receptors, JAKs and STATs still remains largely elusive duo to the lack of structural information. In vertebrates, there are many ligands, receptors, JAKs and STATs making this signaling pathway very complex to be analyzed. By contrast, the JAK/STAT pathway in the fruitfly Drosophila melanogaster is less redundant offering a simpler model to study.
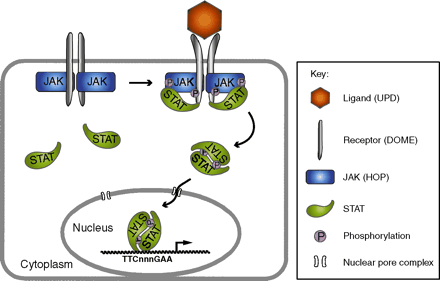
This
diagram is adapted from Development.
133, 2605-16 (2006) [PubMed]
[Full
text]
III. The telomere system of the Streptomyces linear plasmid SCP1
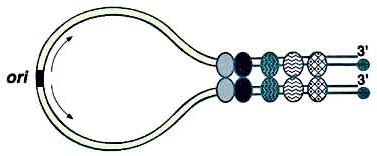
Linear plasmids and chromosomes of Streptomyces carry terminal proteins (TPs) covalently attached to the 5' ends of the DNA. Most known telomeres are conserved in primary sequence and in the potential secondary structures formed during replication. The TP that caps these telomeres is also highly conserved and its coding gene, tpg, is present in most Streptomyces chromosomes and some linear plasmids. However, linear plasmid SCP1 of Streptomyces coelicolor contains atypical telomere sequences and no tpg homologue, and can replicate in the absence of tpg, suggesting that it carries a novel TP gene. A gene on SCP1 (designated tpc) encodes the TP that caps the SCP1 telomeres. The Tpc protein sequence does not resemble the Tpg proteins. A gene (designated tac) upstream of tpc was also required for replication of the SCP1 telomeres. These findings indicate that the Tac-Tpc system of SCP1 represents a novel telomere-capping system of Streptomyces linear replicons.
Racket frame structure proposed for linear Streptomyces plasmids.The black circles represent the terminal protein attached to the 5' ends, which is required to protect the ends and complete the replication of both plasmid termini. The ovals represent juxtaposition proteins that bring together the plasmid termini by binding to specific regions of palindromic symmetry. The ori located near the center of the plasmid is depicted by the box and the arrows indicate the bidirectional DNA replication from this ori.
This diagram is adapted from Front. Biosci.4, D43-62 (1999) [PubMed] [Full text]